By Ron Klauda, Mary Hoover, and Evan Klauda
With apologies to Julie Andrews and “The Sound of Music” for the titular pun, we want to tell you about an exciting biological sampling tool tested by the Friends of Hunting Creek (FOHC) for the first time in Calvert County streams (or rills, if you prefer). Taking inspiration and encouragement from Dr. Chris Rowe (Associate Professor, Chesapeake Biological Lab), we conducted a pilot study using eDNA sampling in four non-tidal streams in the Hunting Creek watershed.
What is eDNA and how can it describe biodiversity?
Environmental DNA, or eDNA, is the genetic material shed by fish and other aquatic animals into the streams, rivers, ponds, lakes, estuaries, wetlands, and oceans where they live. By carefully collecting water samples that contain mucus, skin, scales, other tissues, urine, and yes, even ‘poop’, scientists can extract and process eDNA to learn who lives where.
There is a growing consensus among the scientific community that eDNA analysis is a complementary and perhaps an emerging alternative approach to traditional sampling methods. eDNA analysis is often an easier, quicker, and cheaper way to reliably describe fish communities in a stream. There is also considerable evidence to support using eDNA concentrations as an ancillary tool for estimating fish species composition and abundance, in addition to mere diversity. eDNA-based methods are revolutionizing biodiversity monitoring by enabling non-invasive, efficient, and less costly surveys of diverse taxa in aquatic ecosystems. The FOHC has neither the equipment nor the staff needed to sample stream fishes using traditional sampling gear, so eDNA sampling offered us a doable approach.
To learn more about this innovative sampling tool, check out this short video from “down under.”
What do we want to learn from our pilot study?
Our goal was to determine if eDNA analysis can tell us what fish species call the Hunting Creek watershed home. To better understand and protect the largest watershed in Calvert County (almost 20,000 acres), the FOHC needs to know what lives in the watershed’s 50 or more miles of streams. How many fish species live in these streams and which ones are most common? Are there any rare species in need of special protection? Are there non-native, invasive fish species that could pose a threat to native species? We asked eDNA sampling to help us answer these important questions.
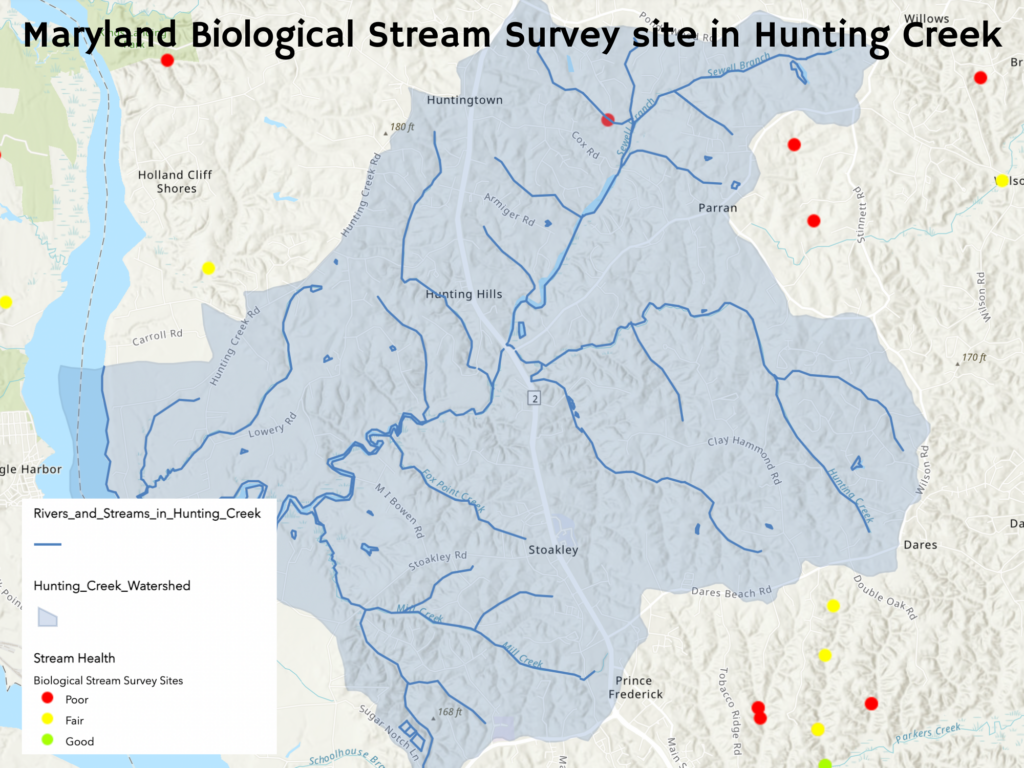
After consulting Maryland DNR’s Stream Health Map, we were surprised to learn there is only one site in the Hunting Creek watershed where fish were collected by the Maryland Biological Stream Survey (MBSS): an unnamed tributary to Sewell Branch that the FOHC unofficially call “Barberry Branch.” Sampled by the MBSS in 2004, only three fish species were collected at this site: Eastern Blacknose Dace, Eastern Mudminnow, and Tessellated Darter. For comparison, the MBSS has sampled and collected fish at 9 sites in the Parkers Creek watershed.
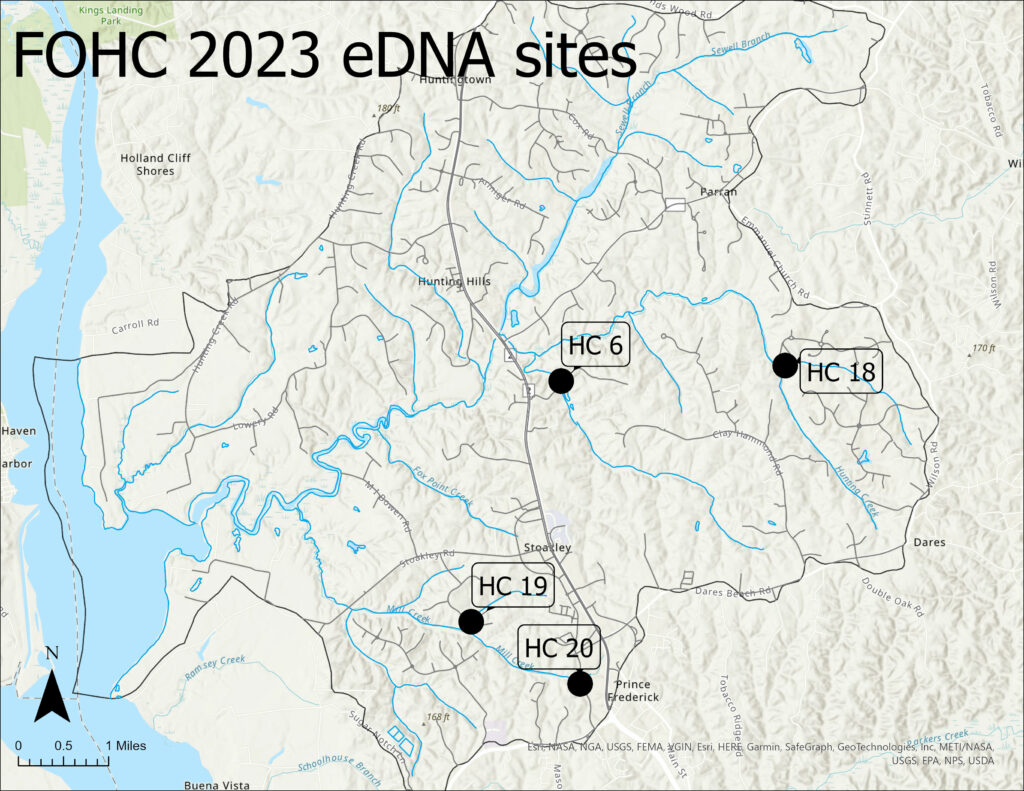
Where, when, and how did we collect eDNA samples?
Water samples were collected in mid-April 2023 in four streams using a Smith-Root eDNA Citizen Scientist sampling pump and their Self-Preserving Filter Packs with micron mesh filters. Two water samples were collected in each stream, along with two distilled water field blanks to check for possible on-site contamination in sample handling.

The four pilot study streams were HC6 (Fox Run), HC18 (unofficially called “Chingaware Run”), HC19 (an unnamed tributary to Mill Creek), and HC20 (upper Mill Creek).
How were the eDNA samples processed?
After collection, the filter packs were mailed to Jonah Ventures for analysis. The DNA was extracted, amplified, sequenced, and then the found sequences were compared to known DNA sequences to determine what fish species were present in each sampled stream.
Allow us to elaborate a bit. As you may recall from that Biology 101 course you took, strands of DNA are made up of four bases: adenine (A), guanine (G), cytosine (C), and thymine (T). Every organism has its own unique pattern of A, G, C, or T sequences. Scientists at Jonah Ventures labeled each base sequence in a process called metabarcoding. DNA sequences in our stream water samples were compared to other sequences in large DNA databases (think library or fingerprint file) to identify individual fish species.
Rob Aguilar (Research Technician, Smithsonian Environmental Research Center) kindly ‘tweaked’ (our word, not his) the spreadsheet we received from Jonah Ventures, using SERC’s private reference sequence library (CBBI: Chesapeake Bay Barcode Initiative), and improved our confidence in the species identifications.
What have we learned so far?
1.Well, for starters, we now know how to collect uncontaminated water samples from streams for eDNA analysis. Jonah Ventures found no DNA in our two field blanks.
2.We also now know what fish species live in the four streams we sampled. For brevity’s sake, this article will share only what we learned about Fox Run (HC6). eDNA results for the other three streams sampled in our pilot study will be discussed in the FOHC’s 2023 water quality monitoring report currently being prepared.

eDNA analysis told us that Fox Run is home-sweet-home to at least the following 16 fish species, in alphabetical order:
American Eel, Bluegill, Brown Bullhead, Chain Pickerel, or Redfin Pickerel. Creek Chubsucker, Eastern Blacknose Dace, Eastern Mosquitofish, Eastern Mudminnow, Golden Shiner, Green Sunfish, Largemouth Bass, Redbreast, Sunfish or Pumpkinseed, Satinfin Shiner, Spottail Shiner, Tessellated Darter, and Yellow Bullhead, more species found by eDNA analysis than in any of the other three streams.
eDNA analysis could not distinguish Chain Pickerel from Redfin Pickerel, or Redbreast Sunfish from Pumpkinseed. All 16 species are native to some portion of Maryland. None are rare, threatened, or endangered. The three most common fishes found in Fox Run were Tessellated Darter (#1), Creek Chubsucker (#2), and Golden Shiner (#3).
Satinfin Shiner and Spottail Shiner are categorized by MD/DNR as intolerant/pollution-sensitive species found only in good quality streams. Spottail Shiner is also a lithophilic spawner, meaning their eggs develop in the cracks and crevices of clean sand, gravel, and cobble substrates. Hence, sediment deposition that buries the developing eggs greatly diminishes their spawning success. Redfin Pickerel is somewhat sensitive to pollution and found only in fair to good quality streams. The rest are more pollution tolerant/less sensitive and found in any quality stream.
3. Perhaps most important for this Pilot Study, eDNA analysis did not find any “oddball” fish species (e.g., Sockeye Salmon) that should not live in the Hunting Creek watershed. Our results from four streams suggest that the ‘DNA fingerprint files’ for Coastal Plain Maryland stream fish are robust and reliable.
How does fish diversity in Fox Run compare to other Calvert County streams?
The short answer is, “The highest!” Compared to all other County streams that were sampled by the MBSS, Fox Run has the most fish species (16). Coming in second, to date, is a site in Lyons Creek (sampled in 1997) with 15 species, including 5 species that eDNA analysis did not find in Fox Run: Fallfish, Least Brook Lamprey, Rosyside Dace, Tadpole Madtom, and Yellow Perch.
In case you’re wondering, 14 fish species have been collected among 9 sites in 5 Parkers Creek tributaries, including a non-native species: Northern Snakehead.
What’s Next?
Fox Run flows north behind the Fox Run Shopping Center and Calvert Health Medical Center, areas with many acres of paved parking lots, roadways, and rooftops. These non-absorbing (impervious) surfaces make stormwater management very challenging. Many studies have shown that impervious surfaces can be “stream killers.” Further development is being planned in the upper portions of the Fox Run watershed (e.g., Magnolia Ridge residential complex), which could threaten a stream fish community that appears to be the most diverse in Calvert County. The FOHC will continue to be vigilant and work diligently to protect Fox Run and all streams in the Hunting Creek watershed.
In addition to presenting the fish eDNA results for the other three streams included in our pilot study, the FOHC’s 2023 water quality report will also present eDNA results for benthic macroinvertebrates that were collected in the four streams sampled for fish diversity.